Recent updates
2025.10.04.: The custom plot tool can now work with tables containing missing values.
2025.10.03.: The missing permalink bug and the TIFF file format bug have been corrected.
2025.06.25.: Bug corrected: the bug of preventing proper operation in the multigene analysis option of the immunotherapy page has been corrected.
2025.06.16.: Bug corrected: if a variable is used as a filter in the custom analysis page, then the "include in multivariate" option is now automatically unavailable.
2025.06.14.: NEW SUBYSTEM released for clinical data. The analysis is available for breast, lung, colon, and prostate cancer.
2025.03.25.: The selection between percentiles and all available cutoffs is now optional in all analysis pages.
2025.03.15.: The results page has now an updated PDF download option.
2025.02.25.: The "Immunotherapy 2025" dataset has been added to the private version.
2025.02.18.: A bug in the custom plot page was corrected.
2025.01.10.: NEW SUBYSTEM released for breast cancer DNA.
2024.11.06.: A bug in the trichotomization process was corrected.
2024.11.01.: The "Restart" button in the custom plot page now returns to the previously uploaded data table.
2024.10.29.: The custom plot page now displays the time unit correctly in the plot.
2024.10.22.: Percentiles are used now in the best cutoff option to increase analysis speed.
2024.10.12.: A bug of non-working permalink features is now corrected.
2024.09.22.: A new algoritm is introduced for database handling to increase analysis speed.
2024.07.08.: A new security update was implemented to incrase the reliability of the KM-plotter.
2024.06.19.: We have corrected a bug which prevented analysis in certain subsystems.
2024.06.14.: The entire KMplot system was moved to a new server computer. Due to the move there was some downtime, we apologize for any inconvenience.
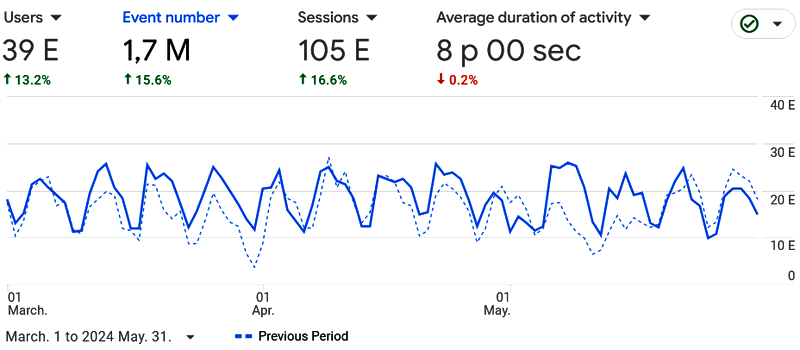
2024.06.01.: We had 39k users who made over one million analyses in the last three months:
2024.04.09.: Our new manuscript describing the colorectal cancer dataset was published in The Innovation.
2024.03.01.: NEW SUBYSTEM released for pancreatic cancer.
2024.02.20.: A bug in the custom plot anylsis page was fixed.
2024.01.28.: Bugs in the Breast protein and Pan-cancer DNA pages were corrected.
2023.12.06.: Stage 1+2, 2+3, 3+4, 1+2+3, and 2+3+4 can now be analyzed in the pan-cancer analysis page.
2023.11.20.: The analysis startup message has been changed from "loading" to "processing" to reflect the fact that all analyses are run in real time and are not loaded with pre-calculated images.
2023.11.07.: Our Google Analytics monitor has been migrated to GA4. We run about 18,000 analyses per day.
2023.09.13.: The colon cancer database has been updated and now has OS data for 1061 patients and PPS data for 311 patients.
2023.09.12.: GSE26906 was removed from the database because this cohort only included redundant samples.
2023.04.18.: NEW SUBYSTEM released for AML.
2023.04.17.: NEW SUBYSTEM released for myeloma.
2023.04.16.: NEW SUBYSTEM released for colon cancer.
2023.04.01.: The lung cancer database was updated and expanded with cohorts published in the last five years.
2023.03.06.: Data table headers can now include the characters "=" and "()" in the custom plot analysis page.
2023.03.05.: Survival time can now be changed after uploading the data file in the custom plot page.
2023.03.01.: Our paper describing the updated ovarian cancer database was published in Geroscience.
2022.10.21.: A bug in the ovarian cancer analysis page was corrected.
2022.08.23.: NEW FEATURE: neoantigen load can now be used as a filter in the pan-cancer analysis page.
2022.08.15.: Due to a network error the server was not accessible for 24 hours. We apologize for the inconvenience.
2022.07.30.: A bug in the immunotherapy subsystem omitting the esophageal adenocarcinoma dataset was corrected.
2022.06.17.: NEW SUBSYSTEM: immunotherapy treated patients from different tumor types including melanoma, bladder cancer, head and neck cancer and available for analysis in the "Immunotherapy" subsystem.
2022.06.01.: NEW FEATURE: the cutoff value can now be manually set in the custom analysis page.
2022.05.28.: Low expression probes are now filtered in the miRNA analysis page and an error message updates the user in such cases.
2022.04.20.: The menu structure of the homepage has been updated and the custom page can now be directly accessed from the top list.
2022.03.02.: A bug of a non-functioning Restart button in some settings has been repaired.
2022.02.28.: A new link to the updated JetSet mapping was added to each array analysis page.
2022.02.25.: Nonfunctional (i) buttons were corrected in each analysis page.
2022.02.20.: The multiple testing link was updated to lead to our new publication for this topic.
2022.02.19.: Bugs in the DNA pan-cancer analysis page were repaired.
2022.01.02.: A missing permalink bug in the results page was repaired.
2021.12.18.: A bug of no results when simultaneously selecting export plot data and trichotomization was corrected.
2021.12.17.: NEW SUBYSTEM: a new analysis platform using RNA-seq data for breast cancer patients was established. The used patient cohort (n=2,976) do now overlap with the samples available in the pan-cancer analysis page.
2021.11.30.: The multiple testing analysis page has been updated to link to our new platform available at www.multipletesting.com.
2021.09.30.: NEW FEATURE: the custom analysis page has been updated and "NA" cells are now permitted in the uploaded table. These cells will be detected and omitted in the multivariate analysis.
2021.08.04.: Out manuscript describing the custom analysis page has been published in the J Med Internet Res.
2021.07.14.: PAM50 subtypes have been added as filtering options to the breast cancer analysis page.
2021.07.05.: A bug related to multivariate analysis in the custom plot analysis page has been corrected.
2021.06.28.: The breast cancer RNA-seq database has been updated.
2021.06.07.: The protein analysis subsystem has been upgraded to enable cross-selection of identical UniProt IDs in case there are different gene symbol used in the individual datasets.
2021.05.20.: A bug affecting settings when using multiple filters in the custom analysis page has been corrected.
2021.04.19.: The option to show the probe expression in normal and tumor tissues has been removed. Please use our new service www.tnmplot.com for this purpose.
2021.04.15.: The Spearman and Pearson correlation p values have been corrected.
2021.03.19.: NEW FEATURE: analysis results can be saved via a new permalink at the end of the results page.
2021.02.20.: The affymetrix breast cancer database has been updated and includes now 7,830 samples.
2021.02.19.: Option to export the generated plot added to the custom plot page.
2021.01.26.: Computation of False Discovery Rate added to the custom plot page.
2021.01.17.: NEW FEATURE: Usage of variables as grouping added to the custom plot page.
2021.01.15.: Proportional hazards computation added to the custom plot page.
2021.01.08.: The background has been improved, URL links have been shortened.
2020.12.08.: The bug of missing beeswarm plot has been fixed.
2020.12.06.: SERVER UPDATE: The auto cutoff is now computed using parallel processes to speed up the analysis.
2020.11.13.: The compute median survival bug has been fixed.
2020.11.05.: NEW FEATURE: A sample file has been added to the custom analysis page.
2020.10.15.: Formatting options have been added to the custom analysis page.
2020.09.28.: Filtering has been extended for ten independent values within variables in custom analysis page.
2020.09.22.: Multivariate analysis has been added to the custom analysis page.
2020.07.22.: New analysis page enabling the analysis of custom data has been established.
2020.06.16.: A bug for correlation analysis in the pan-cancer page has been corrected.
2020.05.25.: In case of 0-0 expression range the system no longer performs an analysis but gives an error message in all tumor types.
2020.05.22.: New icons added to each analysis page.
2020.05.20.: Pan-Cancer: We uncovered a bug due to a file error which resulted in the update we made on the 21st of February. Due to the error some genes were incorrectly indexed in the pan-cancer page, and therefore a result for a different gene was computed for these. As all genes in the first 5000 genes were correctly indexed, none of the quality control runs revealed this problem, this is why it took several months to discover. The file was replaced by a corrected version.
2020.04.15.: The entire KM-plotter was moved to a new faster server computer. In case of IP-address based access, please use 193.6.208.185.
2020.02.21.: NEW FEATURE: Filtering based on cellular content has been added to the pan-cancer analysis subsystem.
2020.02.20.: Updated analysis settings: the best cutoff now selects the highest HR value (or 1/HR in case of HR<1) in case of p=0 in the Cox regression.
2020.01.24.: NEW FEATURE: Use the ratio of genes has been added to the multigene analysis options.
2020.01.23.: NEW FEATURE: An option to filter patients by the expression of another selected gene has been added to the multigene analysis options.
2020.01.02.: NEW FEATURE: The "Plot options" menu now enables to set the legend position and legend font size and type.
2019.12.10.: The maximal number of genes in the multigene analysis was increased to 100.
2019.12.05.: miRNAs with a 0 mean expression were removed from the breast miRNA database.
2019.11.28.: NEW FEATURE: Pearson and Spearman correlation can be computed for any combination of up to four genes.
2019.11.15.: The "Settings" menu has been renamed "Plot options".
2019.11.10.: The "Exclude samples with a protein expression of 0" is now disengaged by default.
2019.11.08.: MAJOR UPDATE: new options for plot formatting: plot line thickness, plot text format, plot text size, plot title and p-value position can now be adjusted in the "More options" menu.
2019.10.25.: NEW FEATURE: Initial release of the proteome-analysis subsystem for breast cancer.
2019.08.01.: A bug of stopped analysis when n=0 has been corrected in the miRpower analysis page.
2019.07.23.: The "remember filtering settings" has been replaced by a new option to "display analysis results in a new window".
2019.07.22.: A bug preventing access to the previous release of the database has been corrected in breast cancer.
2019.06.30.: A new menu option regarding quality control steps implemented in the KM-plotter has been started. The page will be filled with content in the next few months.
2019.05.27.: The bug of incomplete output in case the "each separately" option is selected in the breast cancer dataset selector has been corrected.
2019.04.18.: The esophageal carcinoma dataset has been split to Esophageal Adenocarcinoma and Esophageal Squamous Cell Carcinoma.
2019.04.11.: The lung cancer analysis page has been simplified. Features with minimal usage were moved to the Settings menu.
2019.04.09.: The ovarian cancer analysis page has been compressed to fit into a smaller page.
2019.04.08.: The breast cancer analysis page has been simplified. Features with minimal usage were moved to the Settings menu.
2019.04.01.: RFS, DSS and PFS have been added to the liver cancer RNAseq analysis subsystem.
2019.03.31.: Relapse-free survival has been added to the pan-cancer analysis subsystem.
2019.03.25.: Grade and Mutational burden have been added as filters to the pan-cancer analysis subsystem.
2019.02.20.: Incorrect display of breast cancer quality control dropdown corrected.
2018.12.20.: The homepage has been updated at multiple locations including the download page.
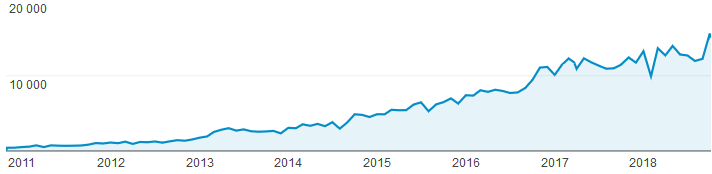
2018.11.04.: Number of visits surpassed the 15.000 limit the first time in October 2018:
2018.11.02.: Structure update for the Pan-Cancer subsystems.
2018.10.09.: Initial release of the PAN-CANCER mRNA subsystem.
2018.10.06.: Initial release of the PAN-CANCER miRNA subsystem.
2018.10.05.: The bug of not running in certain settings was corrected in the miRNA analysis pages.
2018.06.15.: The breast miRNA database has been updated. Both the number of available samples and genes increased.
2018.05.05.: The bug os failing trichotomization in case multiple genes are selected has been corrected.
2018.04.25.: The bug of incorrectly displaying some gene names as dates since the "gene selection dropdown" update has been corrected.
2018.04.18.: NEW FEATURE: False Discovery Rate is now computed whenever the best cutoff is utilized.
2018.03.16.: The bug of disabling the start button when the control code is displayed has been corrected in the breast miRNA analysis subsystem.
2018.03.01.: NEW FEATURE: tirchotomization (T1 vs T3, Q1 vs Q4) added to all cancer types.
2018.02.28.: Post-Progression Survival added to gastric cancer.
2018.02.07.: A bug in the Liver RNAseq analysis system of incorrectly selecting genes has been corrected. We suggest to re-run previous analyses to validet the correct results.
2018.01.24.: The gene selector dropdown menu updated.
2018.01.22.: A bug of not selecting correctly the chemotherapy treated patients in the miRpower breast cancer page has been corrected.
2017.12.08.: Watermark re-added to breast cancer quality control options.
2017.11.30.: The sample number bug in liver RNA-seq page was corrected.
2017.11.17.: Improved homepage design launched.
2017.10.07.: Bugs in the miRpower liver analysis page have been corrected.
2017.10.03.: A subsystem-selection menu has been added to each analysis page.
2017.09.30.: Initial release of the miRpower liver subsystem.
2017.06.15.: Broken links in the download page have been corrected.
2017.05.15.: The Export plot data as text feature has been added to the miRNA analysis system.
2017.05.09.: NEW FEATURE: the Affymetrix probe ID is now added to the plot to make saving of the results easier.
2017.04.30.: NEW FEATURE: Backward reference makes it possible to list all datasets providing patients for the performed analysis. Enable the feature in the "Settings" menu.
2017.02.20.: The "Show probe expression in normal tissue:" table now also displays the "sample n" for cancer.
2017.01.13.: NEW FEATURE: it is now possible to Compute the median survival for both cohorts analysed.
2017.01.10.: MAJOR UPDATE: the ovarian cancer database has been updated and includes now 1,816 patients.
2017.01.06.: The "Compute median over entire dataset" has been moved into the "More options" menu.
2017.01.04.: MAJOR UPDATE: the final version of the miRpower breast cancer analysis subsystem has been launched.
2017.01.02.: Adjuvant and neoadjuvant treaments are now available as a separate filters in the breast cancer analysis page.
2016.12.20.: The "Remember filtering settings" option is now extracted from the "Settings" menu to allow swift access.
2016.10.13.: MAJOR UPDATE: the breast cancer database has been updated and includes now 5,143 samples. In addition, the Pietenpol sutypes have been added as a new filter.
2016.08.18.: MAJOR UPDATE: the expression for each gene in normal (non-cancer) tissues is now available in breast cancer.
2016.06.25.: Our manuscript describing the construction of the gastric cancer database has been accepted for publication in Oncotarget. Please see the Download section for more information.
2016.05.02.: Sample numbers on the analysis start pages are now cached in the user's browser. This reduces page load time by 50%.
2016.04.08.: MAJOR UPDATE: the expression for each gene in normal (non-cancer) tissues is now available in lung cancer.
2016.04.17.: Grade combinations are now among the analysis options in ovarian cancer.
2016.04.04.: MAJOR UPDATE: the expression for each gene in normal (non-cancer) tissues is now available in ovarian cancer.
2016.03.15.: MAJOR UPDATE: the expression for each gene in normal (non-cancer) tissues is now available in gastric cancer. To show the expression range (min-Q1-Q2-Q3-max) and the p value of a Mann-Whitney test comparing normal and tumor samples on the results page, tick the "Show probe expression in normal tissue:" checkbox.
2015.02.07.: MAJOR UPDATE: all 54,675 probe sets present on the HGU133 Plus 2.0 array are now available for analysis in gastric cancer.
2016.01.06.: MAJOR UPDATE: all 54,675 probe sets present on the HGU133 Plus 2.0 array are now available for analysis in lung cancer.
2016.01.03.: A bug of replacing the previous gene symbol instead of adding a new symbol in multigene analysis window has been corrected.
2015.12.11.: MAJOR UPDATE: all 54,675 probe sets present on the HGU133 Plus 2.0 array are now available for analysis in ovarian cancer.
2015.11.24.: NEW FEATURE: A multigene classifier ("Use multigene classifier") and the "Each separately" ("Use following dataset") can now be combined to run the mean-based multigene classifier in each dataset separately - available in breast, ovarian and lung cancer.
2015.11.19.: A bug of erroneous HR values for MKI67 in breast cancer multivariate analysis has been corrected.
2015.10.25.: The Download section has been updated to include a table with all JetSet parameters for each of the 54,675 probe sets.
2015.10.10.: NEW FEATURE: "Each separately" added as new option to the dataset selection dropdown menu in breast cancer.
2015.10.05.: Minor update: now the JetSet best probe set is inserted always when entering a list containing multiple gene symbols in the "Use multigene classifier" window.
2015.09.28.: Fixed bug of missing analysis when "each separately" is selected as a filter in lung cancer.
2015.09.20.: Fixed bug of incorrectly displaying the number of patients when the "Remove redundant samples" checkbox is turned off in the "Quality control" menu in breast cancer.
2015.07.01.: NEW FEATURE: HER2 status measured by IHC/FISH is now available as filter for breast cancer (please note: the "intrinsic subtype" filter uses the HER2 expression measured by the gene chips).
2015.06.27.: MAJOR UPDATE: all 54,675 probe sets present on the HGU133 Plus 2.0 array are now available for analysis in breast cancer.
2015.06.27.: A bug of incorrectly displaying outlier arrays has been corrected in the quality control menu.
2015.05.21.: Fixed bug of incorrectly handling probesets with AFFX prefix.
2015.05.01.: The JetSet table has been updated to the v3.0.0 version in all cancer types.
2015.04.30.: A bug of overlapping between the gene selection window and the settings menu has been fixed.
2015.04.16.: Two new publications have been added to the Download section.
2015.02.17.: A new opening menu has been added to the breast cancer analysis page to integrate all quality control parameters (removal of redundant samples, array quality control, proportional hazards assumption, multivariate analysis) at once.
2015.02.15.: NEW FEATURE: multivariate analysis for MKI67 (proliferation), ESR1 (hormone receptor status) and HER2 (ERBB2 receptor status) has been added to the breast cancer analysis page.
2015.02.01.: NEW FEATURE: a test to check the proportional hazards assumption has been added to the breast cancer analysis page.
2015.01.02.: NEW FEATURE: TP53 mutation status can be used as a new filtering parameter in the breast cancer analysis page.
2015.01.01.: NEW FEATURE: In the breast cancer analysis page the computation can now be run using each dataset separately.
2014.11.19.: DATABASE UPDATE: The lung cancer database has been updated with four new datasets. The entire database now includes 2,437 patients. The previous release can be used by setting the "previous release" option to the "2013 version" of the database.
2014.11.02.: DATABASE UPDATE: The ovarian cancer database has been updated with three new datasets. The entire database now includes 1,648 patients. The previous release can be used by setting the "previous release" option to the "2013 version" of the database.
2014.10.29.: NEW FEATURE: In the ovarian cancer analysis page, TP53 mutation status has been added to the filtering options.
2014.10.28.: NEW FEATURE: Array quality control has been deployed to the ovarian cancer analysis page.
2014.10.26.: In the ovarian cancer analysis page, the calculation of the CA125 filter has been changed. Now "Only patients having an average expression of all three probe sets representing CA125 below the lower quartile across all patients are included in the analysis.
2014.10.18.: A bug of missing confidence intervals in case of enabled "Invert HR values below 1" has been corrected.
2014.09.02.: NEW FEATURE: In the ovarian cancer analysis page, an option to run the analysis using each dataset simultaneously has been set up.
2014.08.01.: A bug of not displaying the patient number correctly before the analysis has been corrected.
2014.07.31.: NEW FEATURE: AJCC stage M has been added to the lung cancer filtering options.
2014.07.09.: Due to multiple user requests, the "Censore at threshold:" has been set to be enabled by default.
2014.07.07.: A bug of displaying error notices in the multiple testing page has been corrected.
2014.06.24.: NEW FEATURE: To enable reproduction of the plot in a different software (for example in Excel), the results can now be exported as a text file. To generate the file, tick the "Export plot data as text:" checkbox in the "Settings" menu.
2014.06.13.: NEW FEATURE: The results page has been extended by an option to save the results as a scaleable graphic file. For this, click the "Download plot as a PDF" link below the analysis results.
2014.06.01.: A bug coming when using the "all probe sets per gene" radio button has been corrected.
2014.05.05.: A bug in the gene selection dropdown list has been corrected.
2014.04.10.: A powerpoint template has been added to the download section to enable changing font and text size.
2014.04.04.: A bug in the multiple testing correction page disabling computation of FDR and q value has been corrected.
2014.03.27.: A bug disabling the multigene classifier in certain settings has been corrected.
2014.03.21.: The maximal number of probe sets in a multigene signature has been increased to 65 genes.
2014.03.04.: NEW SERVER: we moved the entire KM-plotter to a new server computer. The new system has twice as many processors and RAM and is therefore a magnitude faster than the old one. However, because of re-installing the system, some bugs and broken links might remain in the pages - please report these so that we can correct the pages. We apologize for any inconvenience.
2014.02.03.: NEW FEATURE: the "Use multigene classifier" has been replaced by a more complex system. Genes can now be inverted and have different weights. Expression of the gene is multiplied by the weight, thus genes with higher weight will have a more profound effect on the average of the signature.
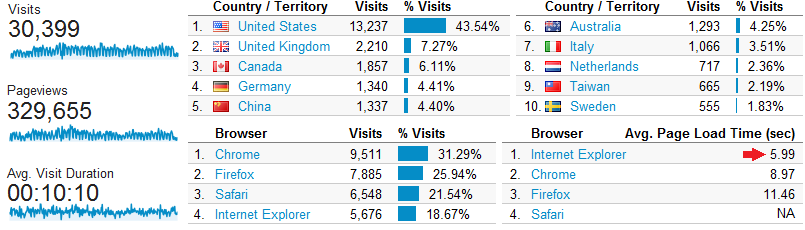
2014.01.10.: Below the Google Analytics summary of visitor activity for http://www.kmplot.com between January 1- December 31, 2013. We suggest to try Internet Explorer in case the page load time is too long.
2013.12.18.: Our paper describing the lung cancer analysis page, multivariate analysis and plotting of funnel plots has been published in PLoS One. Download the paper here.
2013.11.27.: The breast cancer database has been updated and includes now 4,142 patients.
2013.10.27.: Molecular subtypes have been converted to "intrinsic subtypes" to reflect the 2013 StGallen categories.
2013.10.24.: Our manuscript describing the lung cancer analysis page has been accepted for publication in PLoS One.
2013.09.12.: Quick links leading to our related publications have been added to the analysis pages.
2013.09.09.: An option to simultaneously run the analysis for each dataset separately has been added to the lung cancer analysis page.
2013.08.10.: The download section has been updated with our 2013 Breast Cancer Res Treat paper, and R scripts for Kaplan-Meier and ROC analysis.
2013.08.09.: Bug in ovarian cancer analysis page for taxol treated patients repaired.
2013.08.08.: Filtering menu updated in breast cancer analysis page.
2013.07.26.: NEW FEATURE: The "Follow up threshold" menu has been extended by an option to select between censoring and exclusion of patients above the selected cutoff time.
2013.07.24.: NEW FEATURE: An option to compute Post Progression Survival has been added to the KM-plotter. PPS data is available for patients who have relapsed and we have their RFS and OS times as well.
2013.05.22.: The KM-plotter server has been upgraded from a 100Mbps to a 25Gbps permanent internet connection.
2013.05.20.: The follow-up threshold filter has been changed: patients surviving longer than the selected threshold are now censored instead of completely excluded.
2013.04.07.: Initial release of the lung cancer analysis page of the KM-plotter.
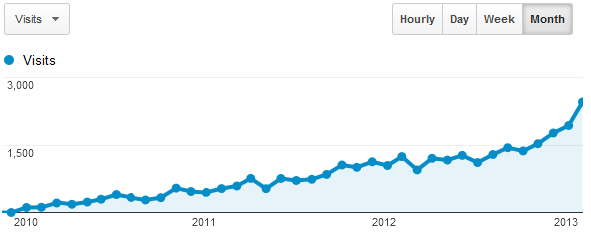
2013.04.04.: Distribution of monthly kmplot.com visits since 2010 (Google analytics screenshot):
2013.03.16.: A performance bug has been fixed.
2013.02.26.: Fixed bug of including non-tamoxifen treated patients in the tamoxifen-only analysis.
2013.02.10.: NEW FEATURE: "tamoxifen only" added to the restriction options in the breast cancer section. The total number of patients with relapse-free survival data in this cohort is 665.
2013.01.31.: Fixed bug of not displaying correctly the number of patients included in the analysis when selecting certain filtering combinations.
2012.11.24.: NEW FEATURE: The breast cancer section has been extended with an option to filter for the array quality control. For more information about array quality control please see our publication Breast Cancer Res Treat 2012.
2012.11.20.: DOWNLOAD SECTION: The download page has been extended with additional documents including our recent presentations about the KM-plotter.
2012.11.08.: DATABASE UPDATE: New datasets have been added to the ovarian cancer database. The analysis now runs on 1,464 samples. The previous release can be used by setting the "previous release" option to the "2012 version" of the database.
2012.10.10.: NEW FEATURES: The treatment-based filtering options for breast cancer have been extended with detailed selection options for systemically untreated/endocrine/chemotherapy treated patients.
2012.10.05.: DATABASE UPDATE: All breast cancer patients included in the restric analysis to selected cohorts have been re-assessed.
2012.06.23.: NEW FEATURE: An option to auto select the best cutoff has been added to the "Split patients" menu.
2012.06.12.: The Enter more Affy IDs box has been re-engineered. The "Use mean expression of the selected probes" ticbox and the "Apply Bonferroni multiple-testing correction" tickbox have been moved here.
2012.06.01.: NEW FEATURE: Breast cancer: an option to use an earlier release of the database has been added to the filtering options.
2012.05.01.: Breast cancer: IHC based PR status has been added to the filtering options.
2012.04.30.: Breast cancer: The molecular subtypes have been changed to use the StGallen criteria.
2012.04.27.: The breast cancer database has been updated with 4 new datasets: GSE16446, GSE17907, GSE19615 and GSE20685. Additionally, redundant samples were removed (the total number of discovered redundantly publised microarrays in GEO is 1418). The analysis now uses the remaining 2997 unique breast cancer samples.
2012.03.22.: Several users requested an option to invert HR values below one. A checkbox for this was added to the Settings menu.
2012.03.09.: The ovarian cancer database has been updated. The analysis now runs on 1,346 samples.
2012.02.03.: The control code has been upgraded to pop-up randomly after a set of analyses were performed (thus the user has not to enter it each time an analysis is run).
2012.02.02.: The output window has been extended to show all gene symbols.
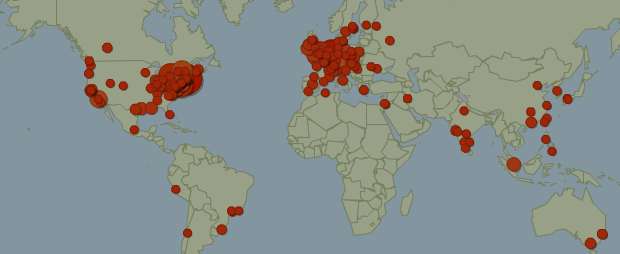
2012.01.21.: Number of visits in 2011: 8,089. Geographical distribution of the users by city:
2012.01.20.: Technical note: a user can run several probe sets simultaneously when using a set of gene IDs AND can also save the analysis settings using the appropriate checkbox in the Settings menu. This way a human user can easily run several analyses without the need to repeatedly enter the control code.
2012.01.10.: The ovarian cancer analysis page has been updated with an option to run single datasets separately.
2011.12.12.: We had several server breakdowns due to thousands of automated analyses run by bots in the last few days. Therefore we have built-in a human recognition control. By using a single character-code only we tried to minimize the load on regular users.
2011.11.28.: A bug of the latest release related to reversed classification of ER status and lymph node status has been corrected. Please re-calculate all ER-status related correlations for analyses made between 14.11.2011 and 28.11.2011.
2011.11.18.: A table containing the exact probe sequences has been added to the background page.
2011.11.14.: NEW FEATURE: The settings menu has been extended with an option to prepare the output graph in TIFF format. Additionally, returning users can mark the latest updates.
2011.11.07.: The settings menu has been extende with an option to save the analysis settings.
2011.11.05.: The analytical page has been extended with a settings menu. For better overview, we have moved some of the minor options into this menu.
2011.10.25.: NEW FEATURE: The probe sets can now be be restricted to JetSet selected best probe sets.
2011.09.05.: NEW FEATURE: The breast cancer analytical page has been extended with the molecular subtypes. The analysis can now be restricted to basal, lumina A, luminal B and HER2? samples.
2011.09.04.: The ovarian cancer analytical page has been extended with the combination of different stage patients. Additionally, the histology groups serous and serous/papillary have been merged.
2011.09.01.: The "Split patients by:" menu has been extended with the option of splitting the patients in lower and upper tertiles.
2011.09.01.: Initial release of the KM plotter for ovarian cancer.
2011.08.05.: Graphical upgrade was performed on the homepage.
2011.08.01.: Initial release of the Multiple-Testing Correction page.
2011.06.11.: The descrtiptions have been updated.
2011.06.06.: DATABASE UPDATE: Another GEO dataset (GSE21653) has been added to the database. The analysis now runs on 2,472 samples.
2011.06.03.: DATABASE UPDATE: Another GEO dataset (GSE17705) has been added to the database.
2011.05.16.: NEW FEATURE: Use multiple probe sets for the analysis. To activate the feature, please use the "Enter more Affy IDs" button. To use the mean expression of the entered probe sets, tick the "Use mean expression of the selected probes" box.
2011.05.16.: Feedback update: the number of patients included in the final analysis are now computed and displayed in real time on the right side. Additionally, the box has a background color representing the robustness of the sample number.
2011.03.28.: NEW FEATURE: Plot beeswarm graph of probe distribution. The bee swarm plot is capable to visualize gene expression as non-overlapping points in a one-dimensional scatter-plot. A beeswarm plot can be used to quickly identify outlier samples.
2011.03.04.: NEW FEATURE: Follow-up threshold. Patients over the selected threshold will not be included in the analysis. This feature is extremely useful for those searching for biomarkers affecting short-time RFS.
2011.01.12.: We had several power failures in the winter holidays. The database engine of kmplot.com has been re-designed to completely restart after power failure.
2011.01.10.: The introduction has been updated.
2010.12.13.: The gene symbol table has been updated using the database published by the HUGO Gene Nomenclature Committee (EMBL Hinxton). Now 70,632 symbols can be used.
2010.11.02.: Gene symbol has been added to the output window.
2010.10.24.: COLOR CODE UPDATE: The color codes for the probe set quality estimation were updated. The original color codes were computed using 1,809 samples, the updated color codes are based on 12,114 samples from 85 GEO datasets.
2010.10.01.: DATABASE UPDATE: Another GEO dataset (GSE2603) has been added to the database. The analysis now runs on 1908 samples.
2010.10.: Our paper desribing the KM plotter has been published in the October issue of Breast Cancer Research and Treatment.